Mission of Laboratory
The recent advances in biomedical research have been producing large-scale, ultra-high dimensional, ultra-heterogeneous data. Due to these post-genomic research progresses, our current mission is to create computational strategy for systems biology and medicine towards translational bioinformatics. With this mission, we have been developing computational methods for understanding life as system and applying them to practical issues in medicine and biology. Our activities and contributions since 2002 are summarized as follows:
Gene Network Analysis
We developed a series of computational methods based on Bayesian networks for mining gene networks from microarray gene expression data. We combined the Bayesian network approach with nonparametric regression, where genes are regarded as random variables and the nonparametric regression enables us to capture from linear to nonlinear structures between genes. An information criterion called BNRC is defined for optimizing parameters and network structures. In order to improve the biological accuracy of estimated gene networks, we made a general framework by extending this method so that it can employ genome-wide other biological information such as sequence information on promoter regions, protein-protein interactions, protein-DNA interactions, and subcelluar localization information. For time-course gene expression data, we developed a dynamic Bayesian network method combined with nonparametric regression. Though the problem of finding optimal Bayesian networks is known computationally intractable, we developed an algorithm for searching optimal and suboptimal Bayesian networks in feasible time for small networks. Computational experiments with this search algorithm have provided evidences of the biological rationality of our computational strategy. These computational methods for estimating gene networks were applied for searching drug target pathways. For a given drug, our strategy assumes two kinds of microarray gene expression data: One is a time-course gene expression data for the drug response. The other is a set of gene expression data obtained by knock-downs of several hundreds of carefully selected genes (one knock-down for each microarray measurement). With these gene expression data, our computational method generates gene networks expressed as Bayesian networks that most strongly relate to the mode-of-action of the drug in cells. In collaboration with University of Cambridge, we prepared more than 350 novel gene knock-downs for HUVEC by using siRNA and the fenofibrate (a drug for hyperlipidemia) was used as the drug for investigation. Microarray measurements were conducted for these gene knock-downs and the drug responses in time-course. From these data, we inferred gene networks of 1000 genes by intensively using the supercomputer system at Human Genome Center. We explored these computationally inferred gene networks for searching drug target genes, by focusing on the genes around PPAR-alpha, which is known as the agonist of fenofibrate. Moreover, this strategy was applied to computationally inferred TNF-α HUVEC networks and discovered two hub genes which were recently proven to regulate inflammation and apoptosis in HUVEC.
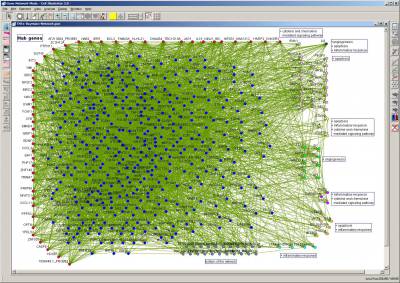 Fig. 1: Gene network computed from the microarray data based on 351 siRNA knock-downs of HUVEC. New hub genes regulating inflammation and apoptosis under TNF-α treatment.
Fig. 1: Gene network computed from the microarray data based on 351 siRNA knock-downs of HUVEC. New hub genes regulating inflammation and apoptosis under TNF-α treatment.
Modeling and Simulation of Biological Systems
We developed a software tool Cell Illustrator™ (CI) with which we can model and simulate various biological mechanisms and pathways in cells, such as metabolic pathways, signal transduction cascades, gene regulations, by organizing and compiling biological data and knowledge. This software is commercialized with a license from University of Tokyo. For this software development, we created a new notion called Hybrid Functional Petri Net with extension (HFPNe) as its architecture. Simultaneously, we have been developing an XML format Cell System Markup Language CSML (http://www.csml.org/) for describing biological systems with dynamics and ontology (Cell System Ontology). The newest version CSML 3.0 covers widely used data formats and applications, e.g. CellML 1.0, SBML 2.0, BioPAX, and Cytoscape. Since CI employs CSML/CSO and equips biology-oriented sophisticated GUIs, we can make modeling of very complex biological processes like with a drawing tool. We used this tool to explore the HUVEC gene networks for the above drug target pathway discovery research.
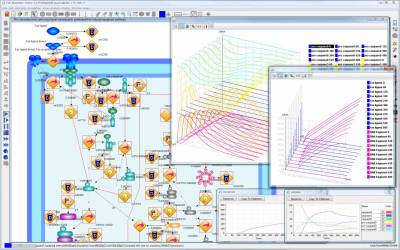 Fig. 2: Cell Illustrator pathway model and its module executing the user specified multiple initial conditions at once and displaying the result with 2D or 3D plots.
Fig. 2: Cell Illustrator pathway model and its module executing the user specified multiple initial conditions at once and displaying the result with 2D or 3D plots.
 Foundations of Systems Biology: Using Cell Illustrator and Pathway Databases (Springer, 2009)
Foundations of Systems Biology: Using Cell Illustrator and Pathway Databases (Springer, 2009)
Peta Flops Computing for Biomedical Research Applications
Since 2006, this laboratory has been involved with the RIKEN’s grand challenge project for life sciences called “The Next-Integrated Life Simulations” which aims at developing software applications that will enable us to simulate and analyze the processes that take place within living organisms, from the molecular level to the level of the whole body. Our key issue is to develop and fuse the data analysis technology and the in silico biosimulation technology. We are challenging this goal with a new statistical and computational technology called “data assimilation” that will realize data-driven computational modeling of biological systems with peta flops computing. As a first step, Cell Illustrator is used to construct a simulatable EGF receptor signal transduction pathway model (EFGR model) based on the biological data and knowledge from the literature. Then, by using some quantitative time-course proteomic data produced with the recent tandem mass spectrometry coupled with liquid chromatography (LC/MS/MS) technology, we semi-automatically constructed a well-tuned EGFR model with the data assimilation technology successfully. For this research direction, we also developed a software tool AYUMS for automatic quantitation of the proteome by LC/MS/MS which shall speed up the analysis process.







