Abstracts
Computational and Verification Platforms for Systems Biology
- P.S. Thiagarajan (National University of Singapore)
 Abstract: We will first present an overview of the research activities of the computational systems biology group in the Dept. of CS at NUS. We will then explore the prospects for exploiting the rapidly advancing GPUs technology for implementing computations relevant for systems biology. In particular, we will outline the potential this technology may have for building large-scale (probabilistic) verification platforms to verify the dynamics of bio-pathways.
Abstract: We will first present an overview of the research activities of the computational systems biology group in the Dept. of CS at NUS. We will then explore the prospects for exploiting the rapidly advancing GPUs technology for implementing computations relevant for systems biology. In particular, we will outline the potential this technology may have for building large-scale (probabilistic) verification platforms to verify the dynamics of bio-pathways.

Incremental Pathway Modeling
- David Hsu (National University of Singapore)
 Abstract: Building quantitative dynamic models of signaling pathways is an important task for computational systems biology. Model construction is an inherently incremental process, with new pathway players and interactions continuously being discovered and additional experimental data being generated. We focus on the problem of performing model parameter estimation incrementally by integrating new experimental data into an existing model. A probabilistic graphical model known as the factor graph is used to represent pathway parameter estimates. A key advantage of our approach is that the factor graph model contains in itself enough information about the old data and uses only new data to refine the parameter estimates without requiring access to the old data. We evaluated our approach by applying it to the Akt-MAPK pathways, which regulate the apoptotic process and are among the most widely studied signaling pathways.
Abstract: Building quantitative dynamic models of signaling pathways is an important task for computational systems biology. Model construction is an inherently incremental process, with new pathway players and interactions continuously being discovered and additional experimental data being generated. We focus on the problem of performing model parameter estimation incrementally by integrating new experimental data into an existing model. A probabilistic graphical model known as the factor graph is used to represent pathway parameter estimates. A key advantage of our approach is that the factor graph model contains in itself enough information about the old data and uses only new data to refine the parameter estimates without requiring access to the old data. We evaluated our approach by applying it to the Akt-MAPK pathways, which regulate the apoptotic process and are among the most widely studied signaling pathways.

Simulation-Based Model Checking Approach to Cell Fate Specification during Caenorhabditis elegans Vulval Development by Hybrid Functional Petri Net with Extension
- Chen Li (University of Tokyo)
Abstract: Model checking is a powerful technique for automatically verifying the requirements of finite state concurrent systems. Since the size of the state space grows exponentially with the number of processes when dealing with continuous values, this problem is generally intractable and serves as a major obstacle hampering further advancement. Till now, researchers generally deal with such problem by means of discretizing discrete model or continuous model of biological networks under specific abstraction criteria. We establish a quantitative methodology by handling quantitative values without such discretization, to model and analyze an in silico model incorporating the use of model checking based on a biosimulation tool Cell Illustrator Online (CIO).
We construct above large-scale quantitative vulval precursor cell fate specification model of C. elegans with CIO. This probabilistic model involves totally 1761 components (place: 426, transition: 442, arc: 780). We performed 480,000 simulations and examined the consistency and correctness of the model under 48 sets of genotypes that are the combinations of four genes and one anchor cell. This method is proved to be a useful means to give researchers valuable biological insights and better understandings of biological systems and observation data that are hard to capture with the qualitative approach.
Next-Generation Cancer Genome Analysis Pipeline
- Tetsuo Shibuya (University of Tokyo)
 Abstract: Next-generation sequencers have become to produce
extraordinary size of data, and moreover the sequencing speed has been
growing almost 10 times faster every year. The analysis of them is
done on massive super computer, and it is very important to
design and develop an analysis pipeline properly on super computers.
In this talk, we will overview our pipeline under
development and related computational topics for a cancer genome project,
where we plan to sequence the genomes of 500 liver cancer patient
using next-generation sequencers.
Abstract: Next-generation sequencers have become to produce
extraordinary size of data, and moreover the sequencing speed has been
growing almost 10 times faster every year. The analysis of them is
done on massive super computer, and it is very important to
design and develop an analysis pipeline properly on super computers.
In this talk, we will overview our pipeline under
development and related computational topics for a cancer genome project,
where we plan to sequence the genomes of 500 liver cancer patient
using next-generation sequencers.
A Novel Meta-Analysis Approach of Cancer Transcriptomes Reveals Prevailing Transcriptional Networks in Cancer Cells
- Atsushi Niida (University of Tokyo)
 Abstract: Although microarray technology has revealed transcriptomic diversities
underlining various cancer phenotypes, transcriptional programs controlling
them have not been well elucidated. To decode transcriptional programs
governing cancer transcriptomes, we have recently developed a computational
method termed EEM, which searches for significantly coherent expression
modules from prescribed gene sets defined by prior biological knowledge like
cis-regulatory elements. In this study, to systematically analyze
transcriptional programs in broad types of cancer, we apply EEM to 122
microarray data sets retrieved from public databases. The data sets contain
about 15000 experiments for tumor samples of various tissue origins
including breast, colon, lung and so on. This EEM based meta-analysis
successfully identified expression modules activated in broad types of
cancer transcriptomes. Furthermore, we predicted transcriptional networks
governing these expression modules as “meta-networks”, which suggest that
cell-cycle and immune related transcriptional programs is employed by
various types of cancer cells. This study demonstrates broad applicability
of our method, and opens a way to comprehensive understanding of
transcriptional networks in cancer cells.
Abstract: Although microarray technology has revealed transcriptomic diversities
underlining various cancer phenotypes, transcriptional programs controlling
them have not been well elucidated. To decode transcriptional programs
governing cancer transcriptomes, we have recently developed a computational
method termed EEM, which searches for significantly coherent expression
modules from prescribed gene sets defined by prior biological knowledge like
cis-regulatory elements. In this study, to systematically analyze
transcriptional programs in broad types of cancer, we apply EEM to 122
microarray data sets retrieved from public databases. The data sets contain
about 15000 experiments for tumor samples of various tissue origins
including breast, colon, lung and so on. This EEM based meta-analysis
successfully identified expression modules activated in broad types of
cancer transcriptomes. Furthermore, we predicted transcriptional networks
governing these expression modules as “meta-networks”, which suggest that
cell-cycle and immune related transcriptional programs is employed by
various types of cancer cells. This study demonstrates broad applicability
of our method, and opens a way to comprehensive understanding of
transcriptional networks in cancer cells.
Computational and Experimental Analysis of the Complement System under Inflammation Conditions
- Liu Bing (National University of Singapore)
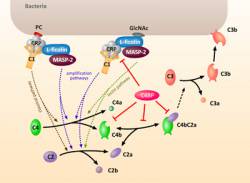 Abstract: Inflammation activates the complement system and induces the crosstalk of the classical pathway and lectin pathway by CRP and L-ficolin interaction. Deficiencies or mutations in the complement proteins will lead to infectious and immune-related disease. How the two pathways balance each other at the system level and the pathophysiological mechanism leading to these diseases are so far unknown.
Abstract: Inflammation activates the complement system and induces the crosstalk of the classical pathway and lectin pathway by CRP and L-ficolin interaction. Deficiencies or mutations in the complement proteins will lead to infectious and immune-related disease. How the two pathways balance each other at the system level and the pathophysiological mechanism leading to these diseases are so far unknown.
In order to quantitatively explain the mechanism of complement system under inflammation condition, we proposed, trained and analyzed a computational model involving the key components of the network. We found that the antimicrobial response is sensitive to the strength of the crosstalk between CRP and L-ficolin which is determined by the pH and calcium level. In addition, inhibitor C4BP plays an important role in maintaining the proper level of complement activation, and this role is implemented mainly by helping the decay of C3 convertase (C4bC2a). These insights are crucial for understanding host defense and pathogen immune evasion, and for the development of complement-immune therapies.
(In collaboration with Prof. Ding Jeak Ling’s Group)

Supercomputing on Gene Network Estimation with Bayesian Networks
- Yoshinori Tamada (University of Tokyo)
 Abstract: The Next-Generation Supercomputer project, led by the Japanese
government, is to develop a 10 peta flops supercomputer by 2012. The
project is not only for building the supercomputer but also for
developing software that can fully utilize the speed of 10 peta flops.
Our research group is being involved in this project and developing
several programs for the gene network analysis. In this talk, I will
present some results of our development on our gene network estimation
software using nonparametric Bayesian networks. Also I will present a
recently developed novel algorithm that realizes genome-wide scale
gene network estimation with nonparametric Bayesian networks.
Abstract: The Next-Generation Supercomputer project, led by the Japanese
government, is to develop a 10 peta flops supercomputer by 2012. The
project is not only for building the supercomputer but also for
developing software that can fully utilize the speed of 10 peta flops.
Our research group is being involved in this project and developing
several programs for the gene network analysis. In this talk, I will
present some results of our development on our gene network estimation
software using nonparametric Bayesian networks. Also I will present a
recently developed novel algorithm that realizes genome-wide scale
gene network estimation with nonparametric Bayesian networks.
Systemic Approach for Finding Differentially Regulated Genes and its Application to Search for Biomarker
- Rui Yamaguchi (University of Tokyo)
 Abstract:
We developed a statisitcal methodology for predicting differentially
regulated genes between case and control samples from time-course gene
expression data by utilizing predictive ability of a state space
model. The proposed method can screen out genes that show different
patterns but generated by the same regulations in both samples, since
these patters can be predicted by the same model. Such kind of genes
are hard to be discriminated by conventional methods. In this talk,
we present a process of the method with an actural example in which
time-course gene expression data from human normal lung cell treated
with(case)/without(control) a drug gefitinib, an inhibitor of
epidermal growth factor receptor tyrosine kinase, are used. We also
discuss an application of the identified differentially regulated
genes as a predictive signature for survival of non small cell lung
cancer patients.
Abstract:
We developed a statisitcal methodology for predicting differentially
regulated genes between case and control samples from time-course gene
expression data by utilizing predictive ability of a state space
model. The proposed method can screen out genes that show different
patterns but generated by the same regulations in both samples, since
these patters can be predicted by the same model. Such kind of genes
are hard to be discriminated by conventional methods. In this talk,
we present a process of the method with an actural example in which
time-course gene expression data from human normal lung cell treated
with(case)/without(control) a drug gefitinib, an inhibitor of
epidermal growth factor receptor tyrosine kinase, are used. We also
discuss an application of the identified differentially regulated
genes as a predictive signature for survival of non small cell lung
cancer patients.
A Decompositional Approach to the Bayesian Approximation of Pathways Dynamics
- Joshua Chin Yen Song (National University of Singapore)
 Abstract: [Liu et al., 2009] constructed a discrete probabilistic model in the form a Dynamic Bayesian Network (DBN) to approximate a system of ODEs. For large pathways models, this technique becomes difficult to manage. Here, we propose a decompositional approach by constructing DBNs for the upstream pathway components and then exploiting these DBNs to approximate the downstream components. We present the technique, a case study
and report some results.
Abstract: [Liu et al., 2009] constructed a discrete probabilistic model in the form a Dynamic Bayesian Network (DBN) to approximate a system of ODEs. For large pathways models, this technique becomes difficult to manage. Here, we propose a decompositional approach by constructing DBNs for the upstream pathway components and then exploiting these DBNs to approximate the downstream components. We present the technique, a case study
and report some results.

Granger Causality in Regulatory Networks: Extensions of Vector Autoregressive Models
- Andre Fujita (RIKEN & University of Tokyo)
 Abstract: Understanding of the mechanisms underlying molecular biological processes are essential to identify and characterize causes of diseases, drug targets and several responses to external stimuli. In order to better understand these processes, statistical models were introduced in the last few years. Granger causality-based models (Vector Autoregressive models) are one of them, presenting well established mathematical interpretations to directionality at the edges of the regulatory network. In this presentation we will describe the concept of Granger causality and explore recent advances and applications in gene networks.
Abstract: Understanding of the mechanisms underlying molecular biological processes are essential to identify and characterize causes of diseases, drug targets and several responses to external stimuli. In order to better understand these processes, statistical models were introduced in the last few years. Granger causality-based models (Vector Autoregressive models) are one of them, presenting well established mathematical interpretations to directionality at the edges of the regulatory network. In this presentation we will describe the concept of Granger causality and explore recent advances and applications in gene networks.
Probabilistic Verification of Biological pathway Dynamics
- Sucheendra Kumar Palinappan (National University of Singapore)
 Abstract: The dynamics of biological systems is usually studied through mathematical models using deterministic approaches such as Ordinary differential equations (ODE) or through stochastic approaches. As the scale at which the systems/models being analyzed get larger there is a need to systematically formalize observations about system behavior and establish the extent to which the model is consistent with observations of interest. Recently there has been a lot of interest in adapting techniques of formal verification esp. model checking for this task. Tools such as PRISM have been used to perform probabilistic model checking of biological systems. The major limitation with these approaches is the state space explosion which limits their use to small models. To address this issue, we propose to build a probabilistic verification framework based on Bayesian Dynamic Models (BDM). The BDM is a stochastic approximation method based on dynamic Bayesian networks developed by members of our group. We will present an LTL (Linear Time Temporal Logic) based model checking framework that has been built for the BDM formalism. We will present how we have tested our approach by verifying the dynamics of the EGF-NGF signaling pathway.
Abstract: The dynamics of biological systems is usually studied through mathematical models using deterministic approaches such as Ordinary differential equations (ODE) or through stochastic approaches. As the scale at which the systems/models being analyzed get larger there is a need to systematically formalize observations about system behavior and establish the extent to which the model is consistent with observations of interest. Recently there has been a lot of interest in adapting techniques of formal verification esp. model checking for this task. Tools such as PRISM have been used to perform probabilistic model checking of biological systems. The major limitation with these approaches is the state space explosion which limits their use to small models. To address this issue, we propose to build a probabilistic verification framework based on Bayesian Dynamic Models (BDM). The BDM is a stochastic approximation method based on dynamic Bayesian networks developed by members of our group. We will present an LTL (Linear Time Temporal Logic) based model checking framework that has been built for the BDM formalism. We will present how we have tested our approach by verifying the dynamics of the EGF-NGF signaling pathway.

Comparison of Estimated Gene Network by State Space Model with Time Course Microarray Data of Different Conditions
- Kaname Kojima (University of Tokyo)
Abstract: We estimate gene networks by state space representation of vector
autoregressive (SSM-VAR) model from time course microarray data of
normal lung cells and lung cells treated by simulating EGF receptors
and dosing an anticancer drug Gefitinib. SSM-VAR can overcome
drawbacks of the vector autoregressive model and state space model;
the assumption of equal time interval and lack of separation ability
of observation and systems noises in the former method and the
assumption of modularity of network structure in the latter method. By
comparing the estimated two gene networks, perturbed genes by the
anticancer drug are identified, whose up- and down-stream genes in the
estimated networks may be related to side effects of the anticancer
drug.
